Carotid artery vessel wall segmentation - Team Wall-E
About
Summary
This model segments the vessel wall of the internal, external and common carotid artery on 3D MRI data. It was developed for the Carotid Artery Vessel Wall Challenge, and achieved the top result.
The segmentation is done in two steps, that ensure anatomically plausible, ring-shaped vessel wall segmentations:
-
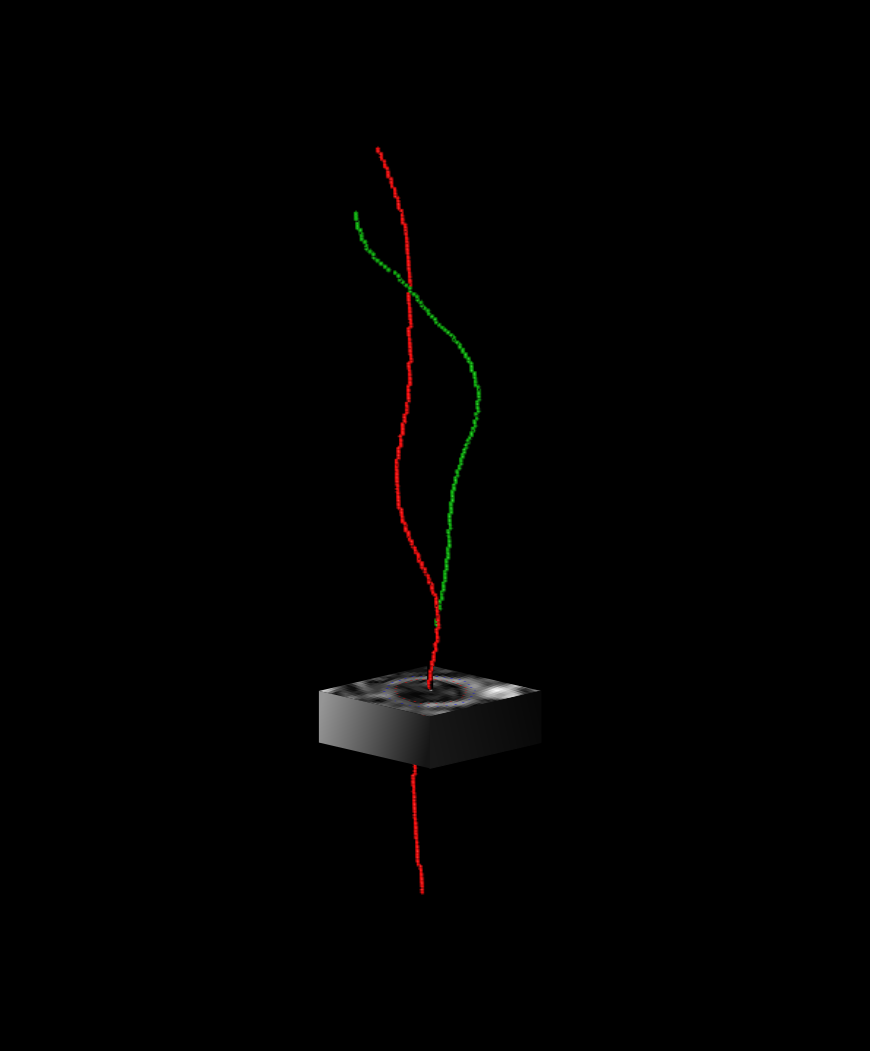
Global: detection of the centerlines of the carotid arteries.
-
Local: vessel wall segmentation around the centerlines, using a polar transform.
The first step uses a 3D U-Net, the second step uses a custom 3D convolutional neural network. The network outputs NumPy arrays, containing contour points of the vessel's lumen and outer wall.
Mechanism
- This method was developed for segmenting atherosclerotic plaque lesions on 3D MRI images of the shoulders and neck.
- The segmentation of the vessel wall happens in two steps, ensuring sub-voxel accurate, ring-shaped segmentations of the vessel wall:
- Global: centerline extraction. We used a 3D U-Net to predict a heatmap describing the proximity to the vessel centerlines. We employ Dijkstra's algorithm to trace the carotid artery centerlines throughout the MRI volume.
- Local: contour estimation. For each axial slice, we extract an image patch centered around the previously found centerline. We perform ray-casting around the center of the image for a set number of angles, resulting in a polar image. This polar image is used as input for a convolutional neural network, that predicts the radius of the vessel lumen and thickness of the vessel wall for each angle. These radii are transformed back to Cartesian space, forming two nested circles, delineating the vessel wall.

The input of the network is a directory containing DICOM images of a 3D black-blood MRI images, similar to the ones available here. On the grand-challenge platform, the outputs are voxel mask of the lumen and wall of each of the 4 carotids (left/right internal/external carotids). To obtain the original outputs (the contours) please contact the authors to access the source code.
Interfaces
This algorithm implements all of the following input-output combinations:
Validation and Performance
| Quantitative score | 0.837 |
|---|---|
| Lumen area difference | 0.086 |
| Wall area difference | 0.072 |
| Hausdorff distance lumen normalized by radius | 0.246 |
| Hausdorff distance wall normalized by radius | 0.215 |
| Dice similarity coefficient | 0.775 |
| Number of unmatched slices | 0 |
All scores were based on the test set of the Carotid Artery Vessel Wall Segmentation Challenge.
Quantitative score is a weighted average of the scores listed below.
Uses and Directions
This algorithm was developed for research purposes only.
Warnings
UserWarning: No point cloud exists for the {side} {label_name} carotid.: This warning may occur if the quality of the found of the annotations was too low to build a segmentation mask for the given carotid. Then the corresponding output will be missing.
Common Error Messages
NoValidSlice: This error occurs when the heatmaps obtained during the first step of the method are too uncertain: then the corresponding carotid is not correctly defined for any of the slices in the volume.
