Autumn Newsletter 2025
Published 12 Dec. 2025
🛠️ New features of Grand Challenge
Welcome to the Autumn Edition of Grand Challenge Insights!
In this blog, we will share the latest updates and highlights from our Grand Challenge community. Following MICCAI 2025, we look back at the outcomes of Grand Challenge-hosted challenges and the contributions of our community during the conference.

Native DICOM Image support now available
We have completed the final features needed for end-to-end DICOM support on Grand Challenge. You can now upload, store, process, and view DICOM images directly on the platform, backed by AWS HealthImaging. This allows algorithms to operate on original DICOM files, aligning more closely with clinical workflows.Customisable end-of-reader-study message
Study organisers can now personalise the message shown to readers upon completing a reader study. Instead of the default “Study finished” popup, organisers can create their own closing message to thank participants, share next steps, or link to a project page.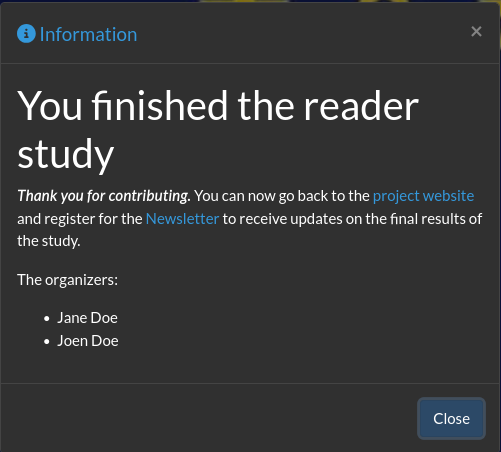

Grand Challenge API client: breaking changes
The Grand Challenge API has been updated to improve consistency and ease of use. Along with the newly implemented DICOM support, several parts of the client were modified to align with these new capabilities. In addition, the documentation has been updated and moved to the client’s own repository for better visibility and maintainability.⚠️ We also used this opportunity to make method and argument naming within GCAPI more consistent across the package, resulting in a number of major changes. Please have a look at the dedicated blogpost regarding these breaking changes.
💡 Blog posts¶
"September 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in September, including continuation of DICOM support and personalised end-of-reader-study messages.
"October 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in October, including steps towards DICOM support and bug fixes for reader study masks.
"November 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in November, expanding on the fully implemented native DICOM support.
🛠️ MICCAI 2025
MICCAI 2025 was an inspiring and productive week for the Grand Challenge team. Two team members attended the conference, where they hosted a workshop and met with many in the research community. They kept notes with feature requests and/or bugs: some of the features have already been rolled out!
In total, around 44% of all MICCAI challenges were organised via the Grand Challenge platform this year, including one of the Lighthouse Challenges (UNICORN), underlining the platform’s continuing role in supporting medical AI research. View all MICCAI challenges hosted on Grand Challenge.
We would like to congratulate all MICCAI Challenge participants for their excellent work and contributions to the field!
MICCAI Lighthouse challenge
🥅 Goal: As part of the MICCAI 2025 Lighthouse Challenges, the UNICORN Challenge established a comprehensive public benchmark to evaluate how foundation models perform across diverse medical imaging tasks. The challenge focuses on assessing their ability to handle the complexity of medical data from multiple domains within a unified framework of 20 tasks.
To enable UNICORN to run on Grand Challenge, new functionalities were implemented. These improvements extend beyond the challenge itself, expanding Grand Challenge’s capabilities for benchmarking foundation models and supporting large-scale, multitask evaluations in the future.
🏆 Winners:
• Best team overall: MEVIS
• Best pathology vision team: MEVIS
• Best radiology vision team: MEVIS
• Best language team: AIMHI-MEDAI
Highlighted MICCAI Challenges
🥅 Goal: The LUNA25 Challenge aims to establish a standardised benchmark for the development and validation of AI algorithms for lung nodule malignancy risk estimation on low-dose chest CT, and to compare their performance with that of radiologists
🏆 Winners:
1. Team Thomas Buddenkotte
2. Team Vaicebine
3. Team MClab
🥅 Goal: PANTHER aims to foster the development of AI solutions that can support clinicians in reducing manual segmentation workload, improving accuracy, and accelerating clinical workflows.
🏆 Winners:
Task 1: Pancreatic Tumor Segmentation in Diagnostic MRIs
1. Team MIC-DKFZ
2. Team SpaceCY
3. Team sjtu_lab426
Task 2: Pancreatic Tumor Segmentation MR-Linac MRIs
1. Team MIC-DKFZ
2. Libo Z. & Team BreizhSeg
🔦 Publication highlight¶
We congratulate the PANORAMA study team on their publication "Artificial intelligence and radiologists in pancreatic cancer detection using standard of care CT scans (PANORAMA): an international, paired, non-inferiority, confirmatory, observational study" in The Lancet Oncology. The study compared radiologists and AI systems for early pancreatic cancer detection on routine CT imaging, and showed AI substantially improved the detection of pancreatic ductal adenocarcinoma. The PANORAMA study was accompanied by a challenge and reader study.
Alves, N., Schuurmans, M., Rutkowski, D., Saha, A., Vendittelli, P., Obuchowski, N., et al. (2025). Artificial intelligence and radiologists in pancreatic cancer detection using standard of care CT scans (PANORAMA): an international, paired, non-inferiority, confirmatory, observational study. The Lancet. Oncology, S1470-2045(25)00567-4. Advance online publication. https://doi.org/10.1016/S1470-2045(25)00567-4
🔦 Highlighted algorithms¶
PTC Segmentation
This algorithm is trained on 59 PAS-stained Whole Slide Images (WSIs) of kidney transplant biopsies to be able to segment peritubular capillaries (PTCs) from the rest of the tissue. By distinguishing PTCs from surrounding tissue structures, this tool can support quantitative morphometric analyses, facilitate large-scale research studies, and potentially aid in diagnostic workflows.
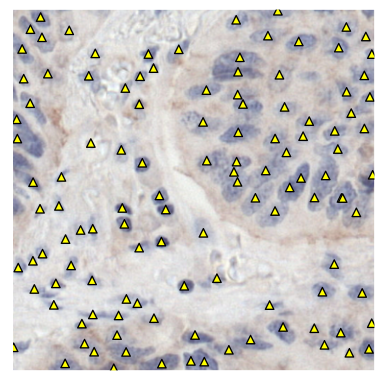
Nuclei detection in immunohistochemistry
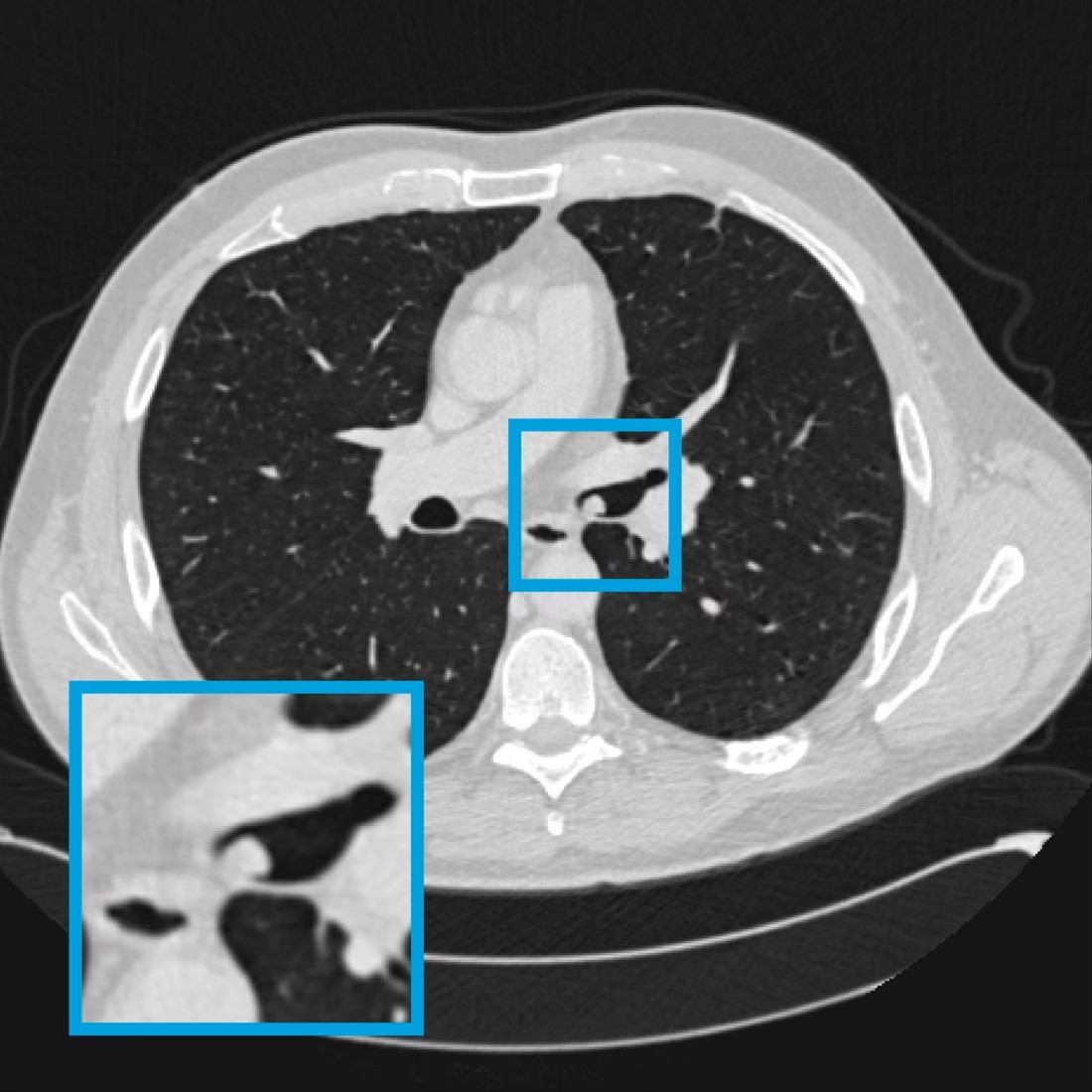
This algorithm automatically detects viable cell nuclei stained with hematoxylin in immunohistochemistry-stained whole-slide images. Originally developed to support semi-automatic annotation, it can also be applied to other tasks requiring precise localisation of nuclei.Airway nodule detection
This deep-learning model detects airway nodules on thoracic or thoracoabdominal CT examinations acquired in routine clinical practice. It is developed on a dataset of 320 CT annotated clinical scans, consisting of 160 scans containing airway nodules and 160 scans without airway nodules.