Hi Z. Huang and Léo Alberge,
Regarding the purpose of different prostate MRI images, and why they can cover a slightly different field-of-view and be misaligned w.r.t. each other, you can refer to this paper:
Regarding the "original" and "resampled" labels:
-
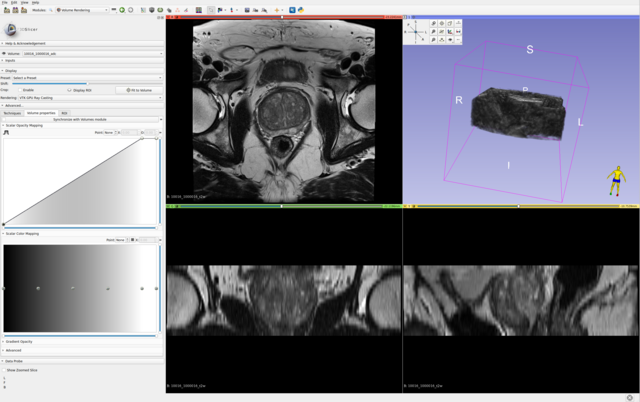
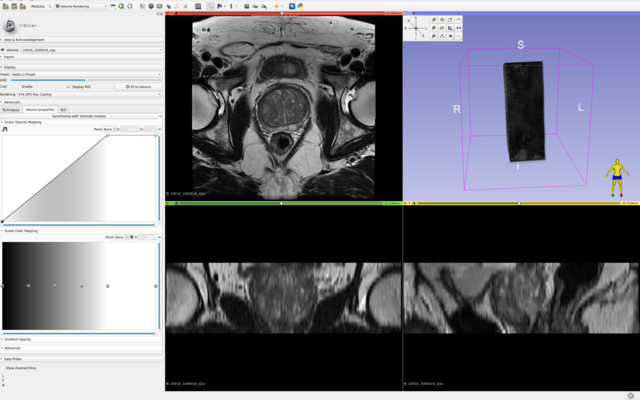
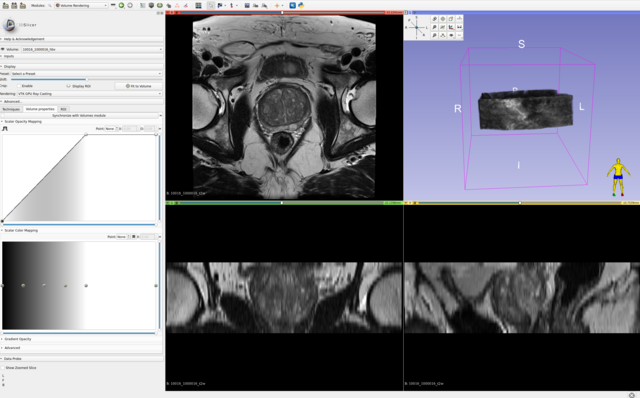
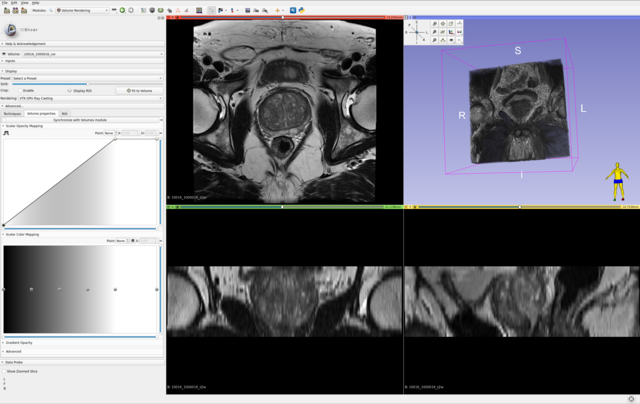
All axial bpMRI sequences (T2W, DWI/HBV, ADC) per case, were used to localize and annotate csPCa lesions. However, depending on the annotator/center and their preference, some annotations have been mapped or created at the spatial resolution of the T2W image, while others have been created at the resolution of the ADC or DWI/HBV images. These original annotations are available in: picai_labels/blob/main/csPCa_lesion_delineations/human_expert/original. For every annotation in this folder, even if the annotation clearly maps to DWI/ADC observations, if it has the properties of T2W imaging, then indeed this annotation was made on T2W imaging (while accounting for observations in DWI/ADC images as well). Similarly, the opposite is also possible. You can determine which sequence was used to segment the tumor(s) for a given study, by looking at the spatial resolution of its annotation file.
-
For a given case, we expect your AI model to predict a csPCa detection map with the same spatial dimensions and resolution as the T2W image. Hence, we have also converted and provided all original annotations at the same dimensions and spatial resolution as their corresponding T2W images, here: picai_labels/blob/main/csPCa_lesion_delineations/human_expert/resampled.
For cases without any substantial inter-sequence misalignment, either of these annotations should be equally valid for all images of the study. For cases with substantial inter-sequence misalignment, annotations will directly correspond to either T2W imaging or DWI/ADC imaging. Note, all cases in the Hidden Validation and Tuning Cohort and Hidden Testing Cohort with any substantial inter-sequence misalignment have been manually co-registered by the organizers. So you shouldn't worry about this at test-time. But for training, this can certainly be something worth exploring.
You can choose to directly use the "resampled" annotations, or preprocess and incorporate the "original" ones, depending on your overall preprocessing and training strategy. Next week, we plan to release picai_baseline: a GitHub repo of baseline AI models that you can use to kickstart your development cycle. Its goal is to help developers get familiar with the end-to-end pipeline of preprocessing prostate bpMRI data, training an AI model for csPCa detection/diagnosis in 3D, and encapsulating the trained AI model in a Docker container for submission to the leaderboard. So you can also refer to those models and their source code, to inform your strategy on which data to use and how to use it.
Hope this helps.