Summer Newsletter 2025
Published 25 Aug. 2025
🛠️ New features of Grand Challenge
Welcome to the Summer Edition of Grand Challenge Insights!
In this blog we will share the latest updates and highlights from our Grand Challenge community.

First steps towards native DICOM support!
We’ve started the first phase of bringing native DICOM support to Grand Challenge. For many years, DICOM files were converted into other formats for use on the platform. Native support will allow algorithms to work directly with DICOM, bringing us closer to clinical workflows. In this initial phase, we’ve developed a de-identification procedure for DICOM tags to help ensure patient anonymity.Optional inputs for algorithms implemented!
We are happy to announce a new feature that allows optional inputs for algorithms. Until now, algorithms on Grand Challenge worked with a fixed set of inputs and outputs. Users trying out the algorithm had to provide all of the defined inputs for the algorithm, and the algorithm had to produce all of the defined outputs. However, inputs often vary in real-world scenarios, and some algorithms can make meaningful predictions with just a subset of inputs. To address this, we've introduced optional inputs, allowing algorithms to handle cases with varying or incomplete data, making the platform more flexible and adaptable.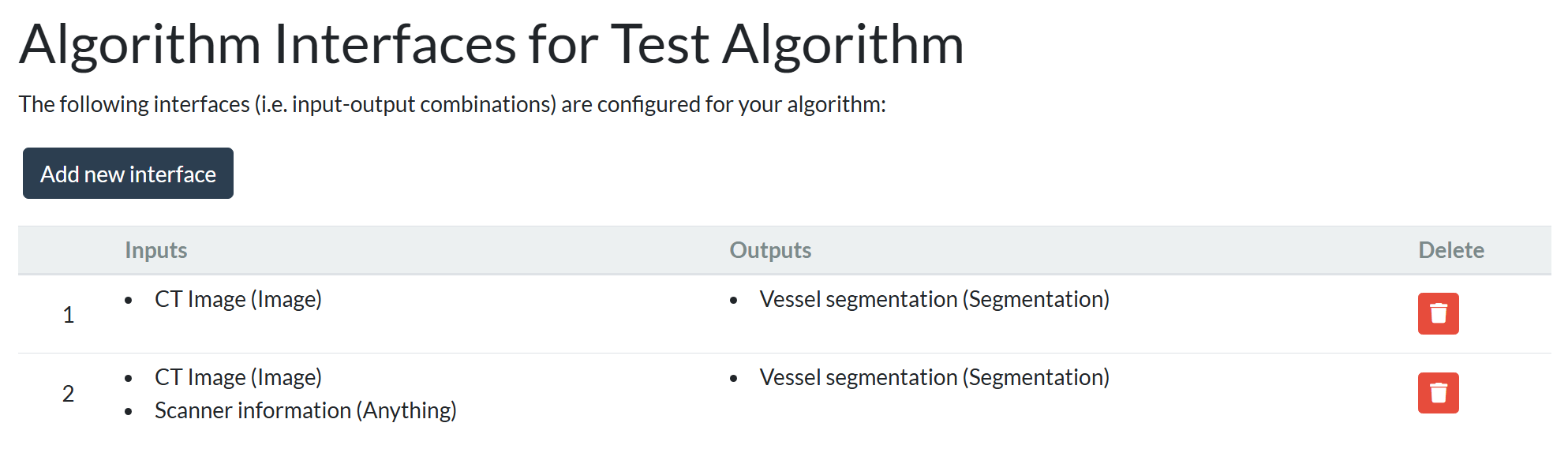
Even more flexibility in evaluating challenge submissions!
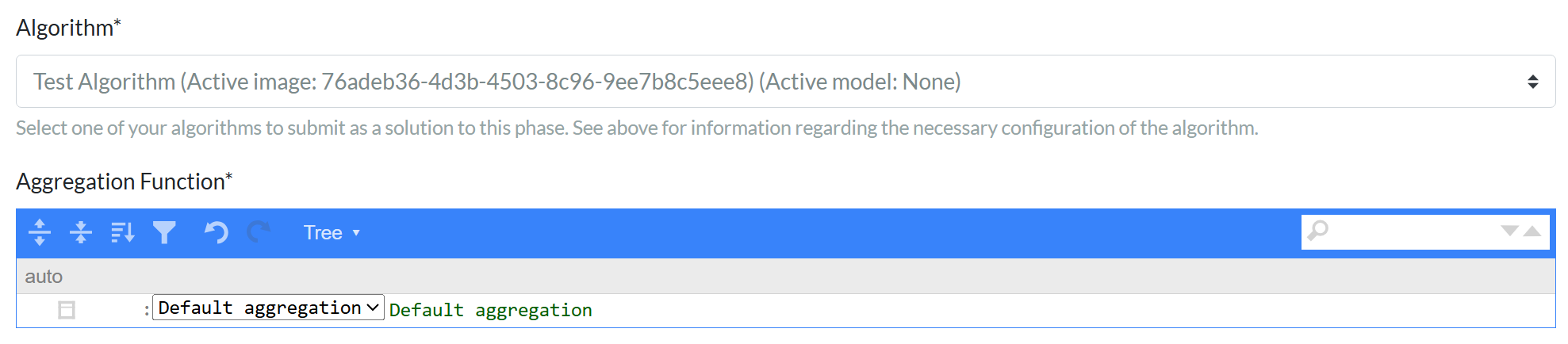
We have also added the option to define extra evaluation inputs, which enables challenge organizers to fine-tune the evaluation with participant-provided options. Challenge organizers can use this to test different evaluation parameters, for example, while reusing the inference results of the algorithms submitted as solutions to the challenge, minimizing time and costs.Challenge task list!
As setting up a challenge can be overwhelming, we have introduced a task list. This task list contains the key steps required to set up a challenge, including estimated time to completion and automated reminders for both the challenge organisers and support staff.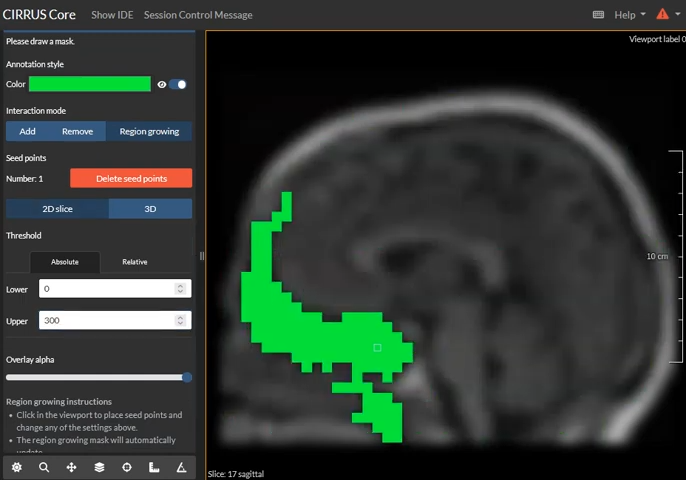
Masks using region growing!
With our newly implemented region growing tool, masks can be quickly generated. Users can set a seed point in the image and a threshold to perform region growing, helping users to quickly and accurately create masks.💡 Blogposts¶
"February 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in January and February, including optional inputs and challenge task lists.
"March 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in March, including new mask-growing tools and an overview of invoices.
"April 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in April, including leaderboards and budget caps for reader studies.
"July 2025 Cycle Report"¶
Read all about the platform improvements from our RSE team in June and July, including the first phase of native DICOM support and auto-saving in reader studies.
🔦 Highlighted algorithms¶
PRISM Embedder
This algorithm extracts slide-level representations from whole-slide images (WSIs) using PRISM. Tile-level features are aggregated using PRISM’s slide encoder to generate a single 1280-dimensional representation for the entire slide. Try out this algorithm with your data!
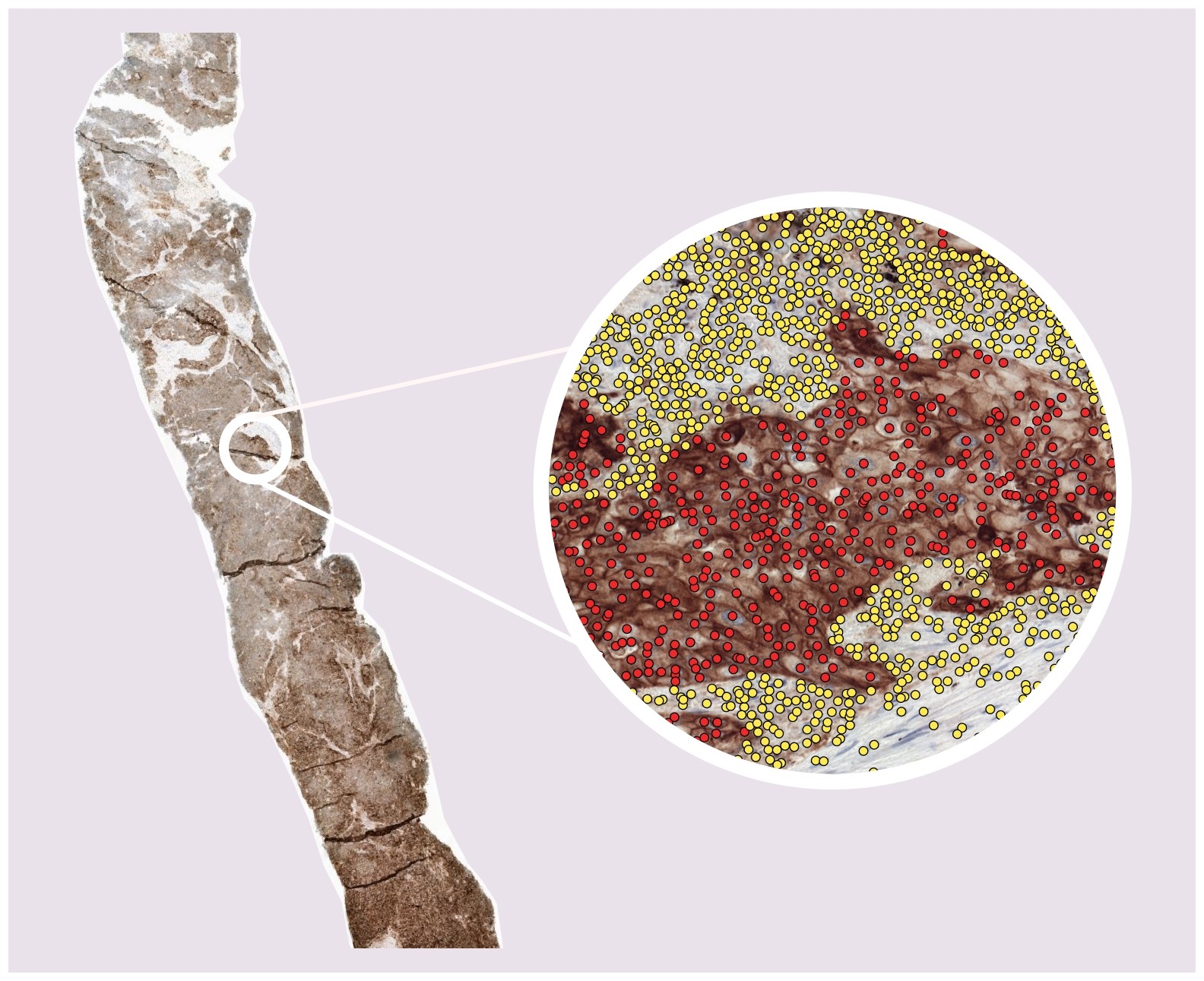
Tumor proportion score in non-small cell lung cancer
This algorithm uses a PD-L1 immunohistochemistry stained whole-slide image to predict the tumor proportion score: the percentage of (viable) PD-L1 positive tumor cells as compared to the total number of (viable) tumor cells in the WSI. Try out this algorithm with your data!🚀 Upcoming and running challenges¶
Challenge activity is at its peak, with many MICCAI challenges now starting or concluding. It's an exciting time for innovation and collaboration across the research community. If you're considering launching your own challenge, you can find all the information you need to get started in our documentation!

🥅 Goal: As part of the MICCAI 2025 Lighthouse Challenges, the UNICORN challenge aims to establish a public benchmark to measure how large-scale generalist models can perform across multiple tasks and handle the complexity of medical data in different domains
among a unified set of 20 tasks.
✍️ Register: Registration is now open!
⏰ Deadline: Accepting AI algorithm submissions to the testing leaderboard until the 29th of August.
🏆 Prize: Prizes will be awarded across four categories:
• Best team overall
• Best pathology vision team
• Best radiology vision team
• Best language team
🥅 Goal: The DRAGON (Diagnostic Report Analysis: General Optimization of NLP) challenge aims to facilitate the development of NLP algorithms, including Large Language Models, for automated dataset curation. The DRAGON challenge includes 28 clinically relevant tasks.
✍️ Register: Registration is open!
⏰ Deadline: The DRAGON challenge will remain open for submissions
🥅 Goal: PANTHER aims to foster the development of AI solutions that can support clinicians in reducing manual segmentation workload, improving accuracy, and accelerating clinical workflows. The challenge consists of two key tasks:
Task 1: Pancreatic Tumor Segmentation in Diagnostic MRIs
Task 2: Pancreatic Tumor Segmentation MR-Linac MRIs
✍️ Register: Registration is now open!
⏰ Deadline: Accepting AI algorithm submissions to the testing leaderboard until the 31st of August.
🏆 Prize: Up to three members from each of the top three performing teams per task will be invited as co-authors.
Task 1:
1. €500
2. €300
3. €200
Task 2:
1. €500
2. €300
3. €200
🥅 Goal: The fourth autoPET challenge explores an interactive human-in-the-loop scenario for lesion segmentation in two tasks: 1) whole-body PET/CT and 2) longitudinal CT.
✍️ Register: Registration is now open!
⏰ Deadline: Accepting AI algorithm submissions to the testing leaderboard until the 1st of September.
🏆 Prize:
Task 1: Single-staging whole-body PET/CT
1. €1500
2. €1000
3. €500
Task 2: Longitudinal CT screening
1. €3000
2. €2000
3. €1000
🥅 Goal: SELMA3D aims to benchmark self-supervised learning for light-sheet microscopy image segmentation tasks.
✍️ Register: Registration is now open!
⏰ Deadline: Accepting AI algorithm submissions to the testing leaderboard until the 15th of September.
🏆 Prize: Top 5 participants will be invited to present their solution and the top 3 will receive Jellycat Selma SLOTH as a souvenir.
🥅 Goal: The RARE 2025 challenge focuses on building a classification system that can accurately detect early-stage cancer in a real-world, low-prevalence setting—specifically, in patients with Barrett’s Esophagus.
✍️ Register: Registration is now open!
⏰ Deadline: Accepting AI algorithm submissions to the testing leaderboard until the 15th of September.
🏆 Prize: All the participating teams will be invited to contribute to the research paper and be listed as authors on the forthcoming journal publication summarizing the outcomes of our challenge.
1. €1000
2. €500
3. €250
4. €150
5. €100
🏆 Leaderboard of finished challenges¶
In the Fuse My Cells challenge, participants had to predict a fused 3D image using only one 3D view, providing a practical solution to the limitations of current microscopy techniques. We would like to extend our heartfelt congratulations to Marek Wodzinski, Shengyan XU, and Cyril Meyer! 🎉
We would like to wish everybody the best of luck with their submissions to the MICCAI 2025 challenges! We look forward to the results presented in Daejeon, South-Korea from the 23rd to the 27th of September 2025.